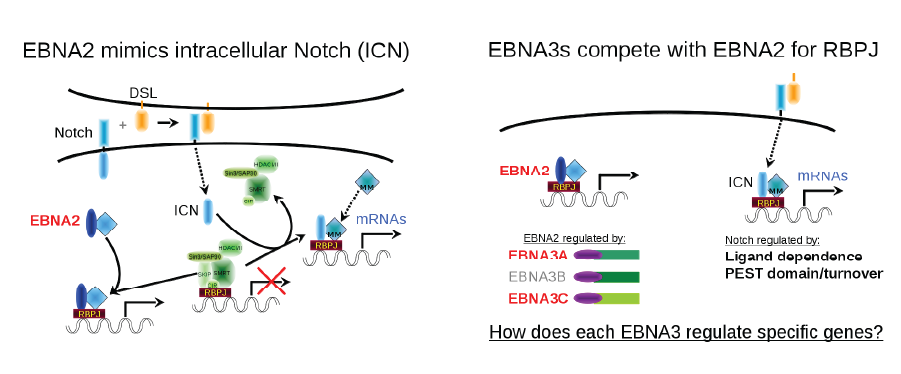
Role of EBNA3 transcription factors in EBV transformation.
EBV Nuclear Antigen 2 (EBNA2) is the master transcriptional regulator of the EBV latency growth program that transforms resting B lymphocytes into lymphoblastoid cell lines (LCLs). During his training, Dr. Johannsen worked in the Kieff laboratory on efforts to identify the cellular factor that targets EBNA2 to promoters. This research showed that this was mediated by interactions with RBPJ, a transcription factor in the Notch signaling pathway. Remarkably, we found that three other EBV nuclear proteins (EBNA3A, EBNA3B, and EBNA3C) also target chromatin via RBPJ interactions. Despite this shared target, each EBNA regulates distinct but partially overlapping sets of host genes through RBPJ and, as subsequent ChIP-seq experiments would suggest, achieve specificity through unique interactions with other host transcription factors.
EBNA2 mimics intracellular Notch (ICN). In the Notch signaling pathway, the intracellular portion of the Notch (ICN) receptor is released upon ligand binding. ICN travels to the nucleus, binds to RBPJ, displacing any repressors that may be complexed with RBPJ and recruiting the activator protein Mastermind (MM), resulting in transcription of Notch regulated genes. EBNA2 mimics this process by directly binding to RBPJ and upregulating expression of EBV latent genes and many cellular genes usually regulated by Notch.
EBNA3A and EBNA3C promote LCL growth by suppressing the expression of the tumor suppressor gene p16INK4A and p14ARF. RBPJ is essential for this activity and EBNA3A or 3C mutants defective for RBPJ interaction fail to suppress p16. Using CRISPR/Cas9, we demonstrated that knockout of p16INK4A was sufficient to restore growth upon EBNA3C inactivation. Our lab maintains an ongoing interest in how the EBNA3 transcription factors reprogram B lymphocytes, including characterization of EBNA3 complexes purified from LCLs. In recent years, the Johannsen Lab has been collaborating with Shannon Kenney’s lab to study the role of EBNA3 proteins in lymphomagenesis using a humanized mouse model.
References:
- Maruo S, Zhao B, Johannsen E, Kieff E, Zou J, Takada K. “Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression.” PNAS 108: 1919-24, 2011. PMCID: PMC3033265
- Wang A, Welch R, Zhao B, Ta T, Keles S, Johannsen E. Epstein-Barr virus Nuclear Antigen 3 (EBNA3) Proteins Regulate EBNA2 Binding to Distinct RBPJ Genomic Sites. J. Virol., 90:2906-19, 2015. PMCID: PMC4810642
- Romero-Masters JC, Ohashi M, Djavadian R, Eichelberg MR, Hayes M, Bristol JA, Ma S, Ranheim EA, Gumperz J, Johannsen EC, Kenney SC. “An EBNA3C-deleted Epstein-Barr virus (EBV) mutant causes B-cell lymphomas with delayed onset in a cord blood-humanized mouse model.” PLoS Pathog. 14(8):e1007221, 2018. PMCID: PMC6117096
- Ohashi M, Hayes M, McChesney K, Johannsen E. “Epstein-Barr virus nuclear antigen 3C (EBNA3C) interacts with the metabolism sensing C-terminal binding protein (CtBP) repressor to upregulate host genes.” PLoS Pathog. 17(3):e1009419. 2021. PMCID: PMC7993866
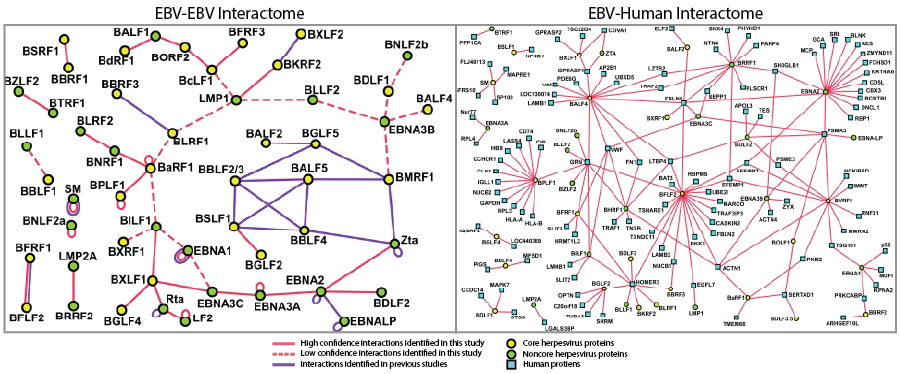
EBV proteomics and interactome map.
Among our most cited manuscripts is a publication that combined classical virion purification techniques with liquid chromatography-mass spectrometry (LC/MS/MS) to analyze the protein composition of the EBV extracellular virion. This analysis revealed that, in addition to EBV encoded proteins, many host proteins are specifically incorporated in these particles. Subsequent efforts, in collaboration with the Vidal lab, to understand the intricate protein-protein interactions involved in this process lead to the publication of the first EBV protein-protein “interactome map.” Many of the interactions identified by this map have formed the basis for additional publications from other labs and my own, including work on LF2.
References:
- Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. “Proteins of purified Epstein-Barr virus.” Proc Natl Acad Sci USA 101:16286-91, 2004. PMCID: PMC528973
- Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, Vidal M, Kieff E, Johannsen E. “Epstein-Barr virus and virus human protein interactome maps.” Proc Natl Acad Sci USA 104:7606-11, 2007. PMCID: PMC1863443
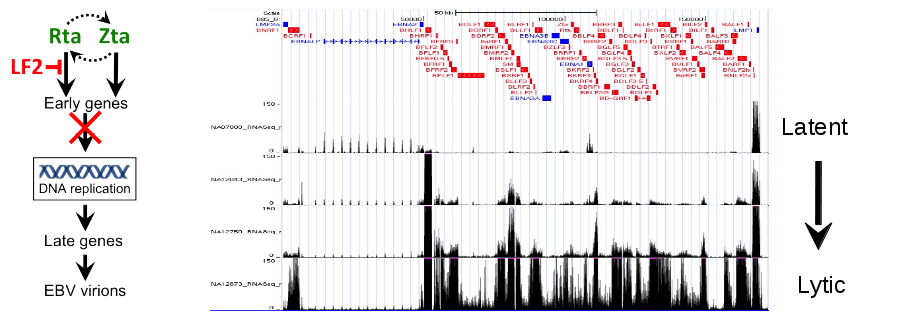
Discovery of LF2 as a regulator of EBV replication.
EBV replication is controlled by two immediate early transactivators, Rta and Zta. In a genome wide screen designed to identify all interactions among EBV proteins we discovered that Rta interacted with LF2, an EBV protein encoded by one of three genes (LF1, LF2, and LF3) present in wildtype EBV, but deleted from the EBV laboratory strain, B95-8. We subsequently discovered that LF2 profoundly inhibited Rta’s ability to upregulate EBV replication genes. In cell-based assays, LF2 can inhibit EBV DNA replication, lytic protein expression, and production of extracellular viral particles (virions), normally induced by Rta. In subsequent studies, we found that LF2 exerts its effects by sequestering Rta in the cytoplasm and preventing it from activating EBV lytic promoters. As part of these studies, a former graduate student, Andreas Heilman, conducted the first genome-wide analysis of Rta binding by ChIP-seq.
LF2 homologs are present in all gamma herpesvirsuses sequenced to date and are incorporated into virions of many of these viruses, including the Kaposi Sarcoma Herpesvirus (KSHV). We speculate that LF2 homologs are incorporated gamma herpesvirus particles to prevent replication upon initial infection, just as alpha and beta herpesviruses incorporate transcriptional activators into their virions to promote it.
References:
- Heilmann AM, Calderwood MA, Johannsen E. “Epstein-Barr virus LF2 protein regulates viral replication by altering Rta subcellular localization.” J. Virol., 84: 9920-31, 2010. PMCID: PMC2937793
- Heilmann AM, Calderwood MA, Portal D, Lu Y, Johannsen E. “Genome-wide analysis of Epstein-Barr virus Rta DNA binding.” J Virol 86:5151-64, 2012. PMCID: PMC3347379
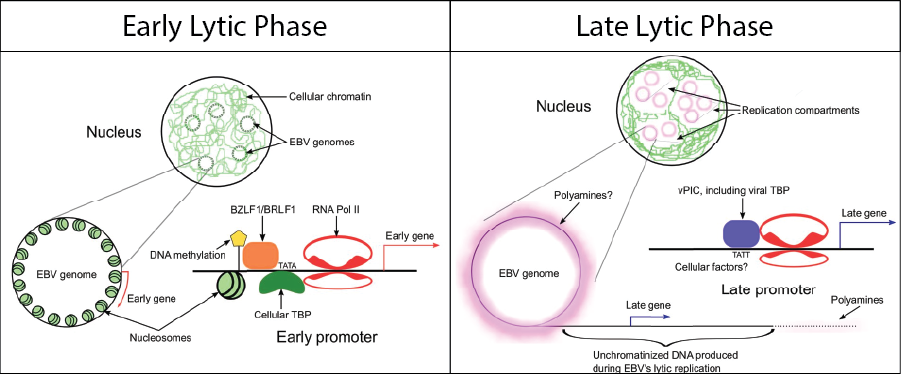
Mechanisms of EBV late gene expression.
Late genes are defined as those that absolutely require viral DNA replication for their expression. Although inhibition of DNA polymerase has long been known to suppress expression of these late genes, the molecular mechanisms linking DNA replication to their expression were unknown. Using EBV recombinant viruses, we showed that 6 EBV genes with homologs found only in beta and gamma herpesviruses (BcRF1, BDLF3.5, BDLF4, BFRF2, BGLF3, and BVLF1) are required for late gene expression. These beta-gamma genes have been investigated by other labs as well and were shown by others to encode a viral pre-initiation complex (vPIC) that recognizes an atypical TATA box (TATT) found in late gene promoters. We found that vPIC can only function if there is an origin of lytic replication present in cis and proposed the model that can explain the dependence of late genes on DNA replication: vPIC serves as an adaptor molecule to recruit RNA polymerase and allow transcription from the newly replicated DNA template. Work by the Sugden lab showed that this newly replicated DNA is unchromatinized and thus vPIC is remarkable in that it subverts the host RNA polymerase to transcribe late genes in the absence of epigenetic signals. We used our EBV recombinants in CAGE-seq experiments to define the dependence of all transcription start sites (TSS) on DNA replication and vPIC. We found that most, but not all late gene TSS require vPIC and demonstrated that some genes appear to be partially dependent upon DNA replication (“leaky”) because they are transcribed by both the early and late gene transcriptional machinery. We recently published a method that improves quantification of lytic gene transcripts from RNA-seq data with accuracy comparable to CAGE-seq.
References:
- Djavadian R, Chiu YF, Johannsen E. “An Epstein-Barr Virus-Encoded Protein Complex Requires an Origin of Lytic Replication In Cis to Mediate Late Gene Transcription.” PLoS Pathog. 12(6):e1005718, 2016. PMCID: PMC4922670
- Djavadian R, Hayes M, Johannsen E. “CAGE-seq analysis of Epstein-Barr virus lytic gene transcription: 3 kinetic classes from 2 mechanisms.” PLoS Pathog.14(6):e1007114, 2018. PMCID: PMC6005644
- Chakravorty A, Sugden B, Johannsen EC. “An Epigenetic Journey: Epstein-Barr Virus Transcribes Chromatinized and Subsequently Unchromatinized Templates during Its Lytic Cycle.” J Virol. 2019 Apr 3. PMCID: PMC6450099
- Casco A, Gupta A, Hayes M, Djavadian R, Ohashi M, Johannsen E. Accurate quantification of overlapping herpesvirus transcripts from RNA-seq data. J Virol. 2021 Oct 27:JVI0163521. PMCID: PMC8791286